So I got around to doing an actual cython performance comparison. As was expected, converting python code to cython results in fairly substantial performance gains. I’ve posted the code on github, and will show the results here. I simply took the primes example in the cython tutorial, and created a python version, and modified the cython version a little bit. The python version is below:
def primes(kmax):
p = [0] * kmax
result = []
k = 0
n = 2
while k < kmax:
i = 0
while i < k and n % p[i] != 0:
i = i + 1
if i == k:
p[k] = n
k = k + 1
result.append(n)
n = n + 1
return result
For the cython version, I modified the original static array in the code to a dynamic array using malloc. This allowed me to pass in the desired length of the primes return, without having to use an upper bound exception, as is done in the original cython tutorial. This was a little frustrating, as I was not familiar with malloc, and it is a good example of why python is such a wonderful language to use (assuming you don’t have performance issues). The cython code is below:
from libc.stdlib cimport malloc, free
def primes(int kmax):
cdef int n, k, i
cdef int *p = <int *>malloc(kmax * sizeof(int))
result = []
k = 0
n = 2
while k < kmax:
i = 0
while i < k and n % p[i] != 0:
i = i + 1
if i == k:
p[k] = n
k = k + 1
result.append(n)
n = n + 1
free(p)
return result
As you can see, there is still some python code in the cython function. One of the limitations with cython, is that if I want to run the cython function from a python script, I need to return the results as a python object. Of course, I could just run everything in cython!
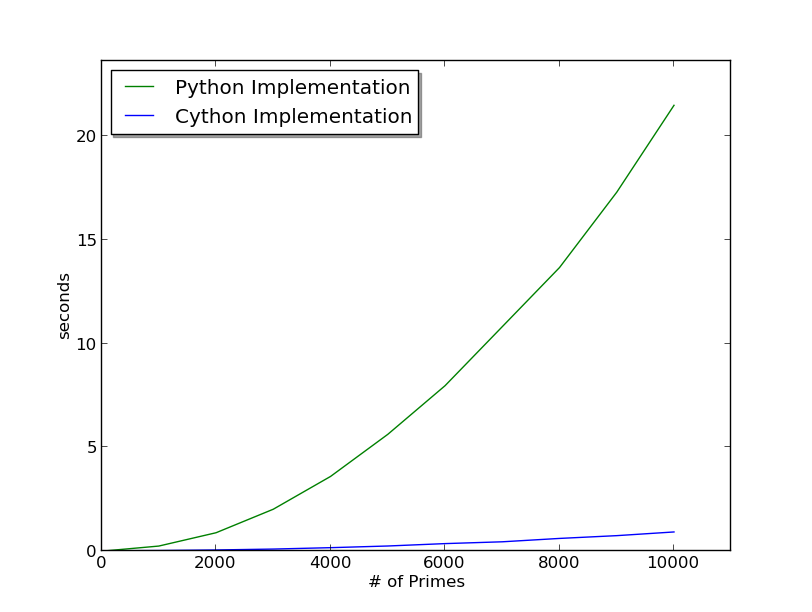
I created a script to time and plot the performance of the two functions over a number of input values. Here is the result:
So as you can see, the partial cython implementation vastly outperforms the python function. This gives you an idea of what kind of overhead the python interpreter brings to the language. So know that I have a good idea of what kind of performance gains I can get using cython, I am going to try to implement some of these in my algorithms. From what I understand, I simply need to convert python data objects to cython data types in order to get the code to compile to C. I think a fun future project would be to write a python script that automatically converts common python data objects to their corresponding cython counterparts. If I get a little time in the near future I may give that a shot.